Comparing representation methods
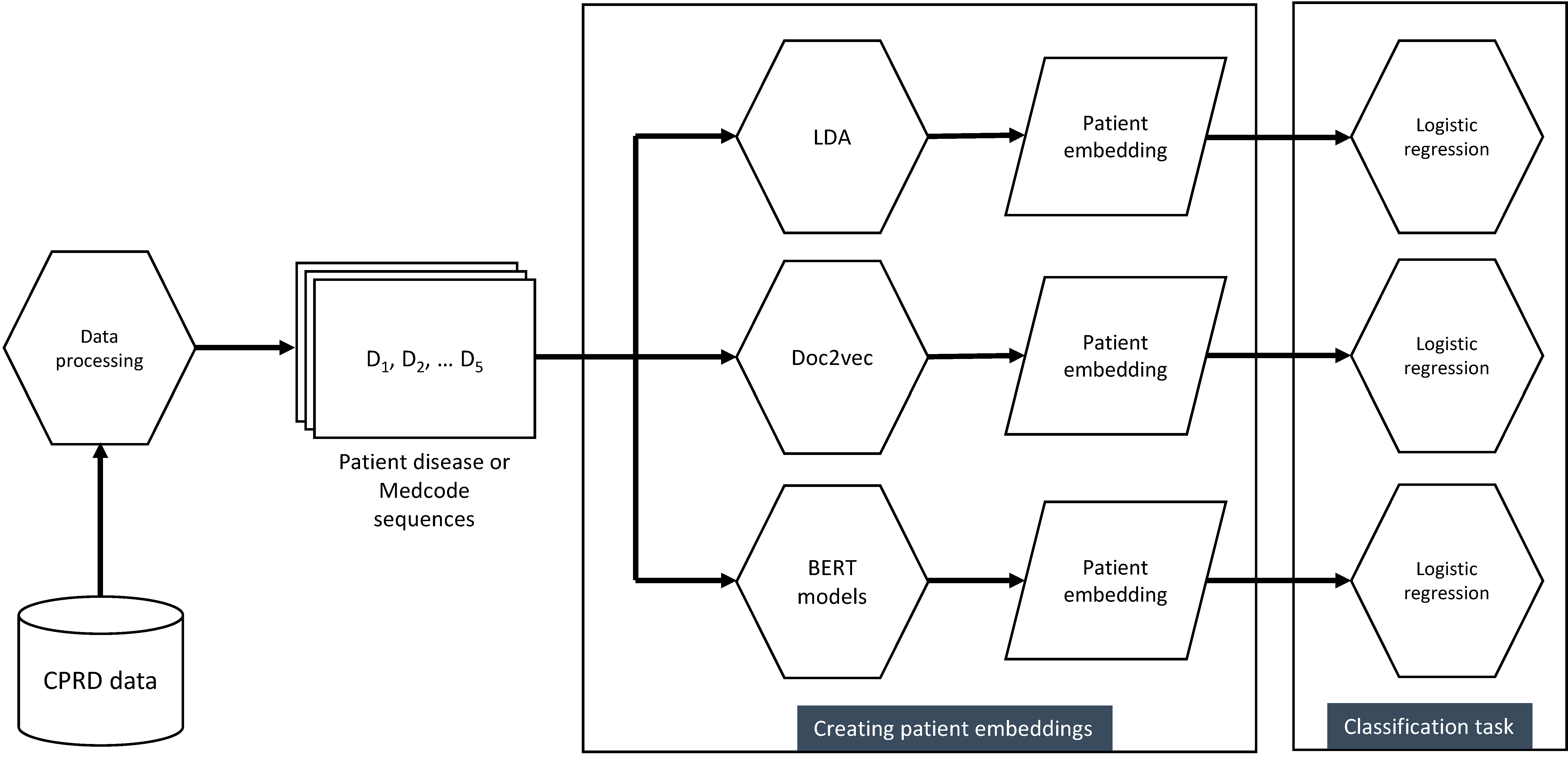
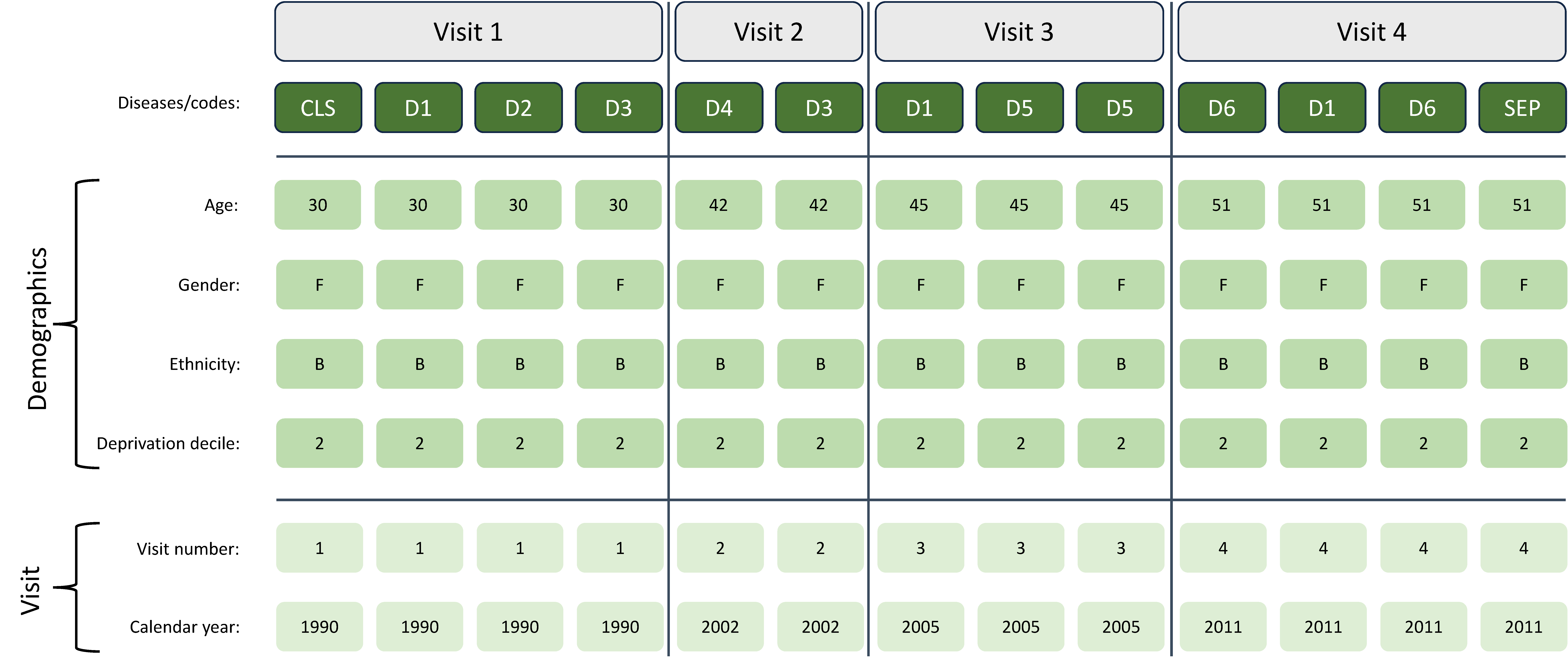
We compared different methods of generating vector representations of people (based on their sequences of diseases), inspired by methods from NLP, including topic modelling and transformer architectures. Each method generates an unsupervised vector representation of 6.3 million people with MLTC. We also compared different inputs of using the raw Medcodes (n=9,462) or disease categories (n=212) as the sequence inputs.
Given the unsupervised generation of these embeddings, we required a strategy to select which embedding to use for clustering. We rationalised that a good embedding will be one the captures a high degree of the inherent information in the disease sequences. Therefore, we developed an evaluation strategy to select the embedding that could best predict future patient outcomes (using data that was not seen in generating the embeddings). We used the patient vectors as inputs into a simple logistic classifier to predict the following outcomes in the 12 months after the embeddings were generated:
- Mortality in those aged ≥ 65 years
- Any emergency department attendance
- Any emergency hospital admission
- Any attendance with a diagnosis code for:
- Hypertension
- Diabetes
- Depression
- A new diagnosis of:
- Hypertension
- Diabetes
- Depression
Analysis pipeline
 Used with permission from Beaney et al (2024)
Used with permission from Beaney et al (2024)
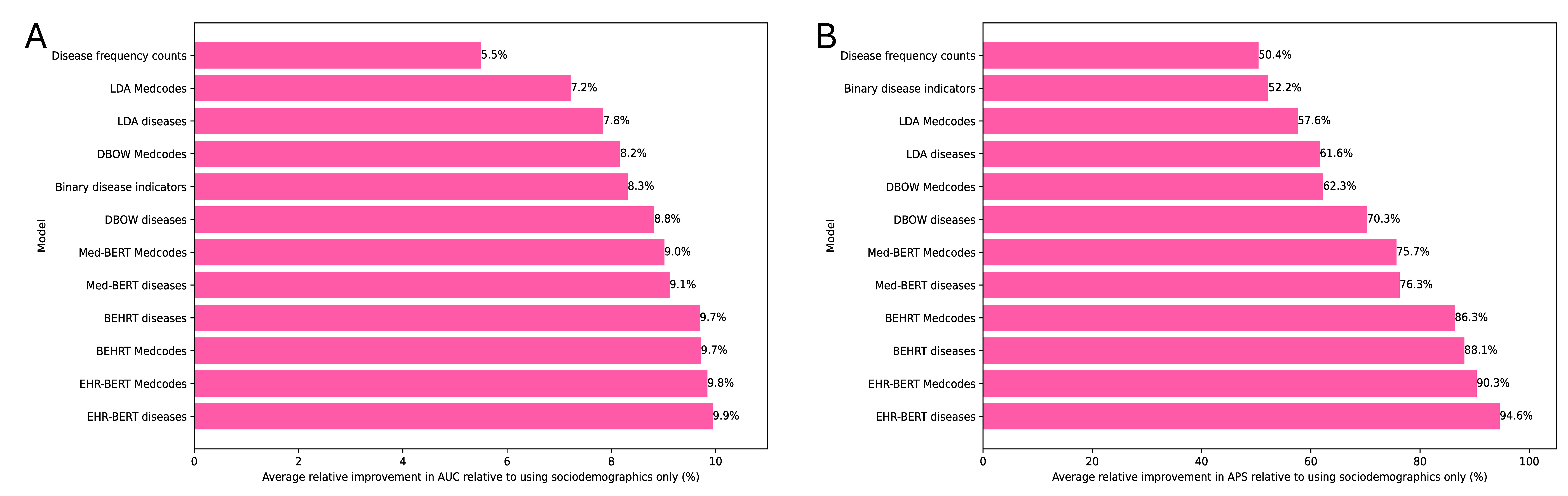
Of the various methods we tested, we found transformer methods to perform best across outcomes. The figure below shows the relative improvement in both AUC and APS averaged across all outcomes for each of the methods that we tested. EHR-BERT was the best-performing method across all outcomes. Use of the smaller number of 212 clinically categorised diseases performed slightly better than use of the larger vocabulary of Medcodes.
Further details of the comparisons and their performance for predicting outcomes are given in our publication in JAMIA.
Embedding performance for prediction
 Used with permission from
Used with permission from